Background
There have been long heated discussions in the literature about the taxonomic status of several subtypes of tick-borne encephalitis virus (TBEV). In particular, it is about the European subtype of TBEV (TBEV-EU). In terms of cladistics, this is due to the fact that the common TBEV clade includes the louping-ill virus (LIV) taxon resulting in paraphyletic relationships between these two closely related species (Fig. 1):
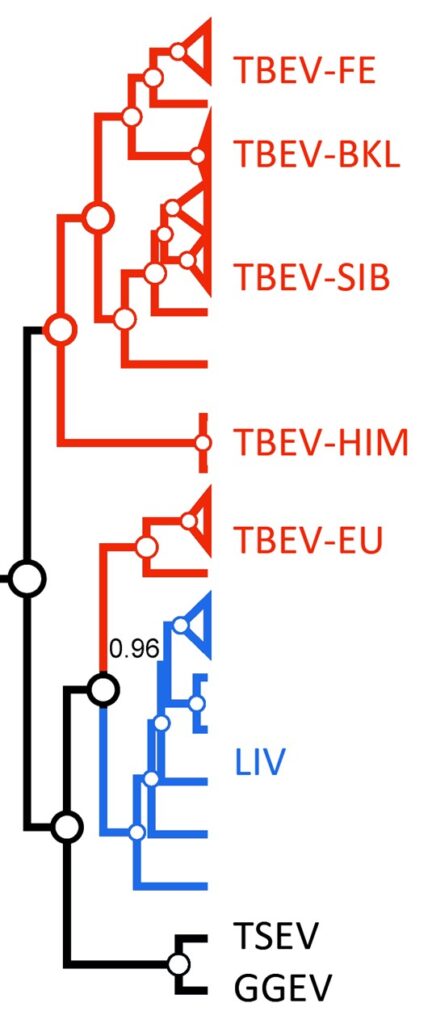
Figure 1: The paraphyly of a TBEV taxon (red color) due to LIV inclusion. White circles depict posterior probability of 1.02. TSEV – Turkish sheep encephalitis virus; GGEV – Greek goat encephalitis virus (both of them are unclassified LIV-like viruses).
In the interim, the species definition claimed by ICTV is “A species is a monophyletic group of viruses whose properties can be distinguished from those of other species by multiple criteria”. To follow this objective principle, TBEV and LIV paraphyletic issue should be resolved. This could be done in two ways: whether by the separation of TBEV-E from the other TBEV subtypes (Far-Eastern [-FE], Siberian [-SIB], Baikalian [BKL], and Himalayan [HIM]) or by merging TBEV+LIV into the single species taxon. The last solution was proposed by Grard et al6 where authors combine TBEV and LIV taxa into the single species based on amino acid (aa) p-distances (0.09) of complete polyprotein aa sequences (4 for TBEV and 4 for LIV). Charrel et al2 came to the same conclusions on the TBEV+LIV integration based, however, on the analysis of E gene sequence (4 for TBEV and 9 for LIV). In both cases, the number of sequences analyzed was small regarding the currently available data. Moreover, the so-called “cut-off rule” (as in the case of Grard et al6) has one major flaw – the absence of biological rationale underlay the threshold proposed. Despite pairwise distance thresholds being able to work well in practice,8 evolutionary methods are needed to validate their use. Also, an underlying evolutionary model makes it possible to compare alternative evolutionary hypotheses statistically.3 Thus, the merger of TBEV+LIV applying only evolutionary distances can be questioned.
Meanwhile, a large amount of data on the eye-catching differences between virus particularities of TBEV-EU and other TBEV subtypes have been accumulated at the moment that enables consideration of TBEV-EU as an independent taxonomic unit.
Thereby, to holistically scrutinize the paraphyletic problem of the TBEV+LIV group, we, at first, employed three available delimitation methods (GMYC, ABGD, PTP) using complete polyprotein aa sequences (n = 278) of all tick-borne flaviviruses (TBFV, 12 species), and finally, we analyzed available literature for other viral species criteria.
Delimitation Analysis
All three delimitation methods demonstrated that TBEV+LIV is not a single species unit. TBEV-E was separated into distinct taxon as well. Some methods such as PTP were tended to fraction TBEV subtypes within the clade more frequently, but none of them merged TBEV+E with other TBEV subtypes or with LIV (Fig. 2):
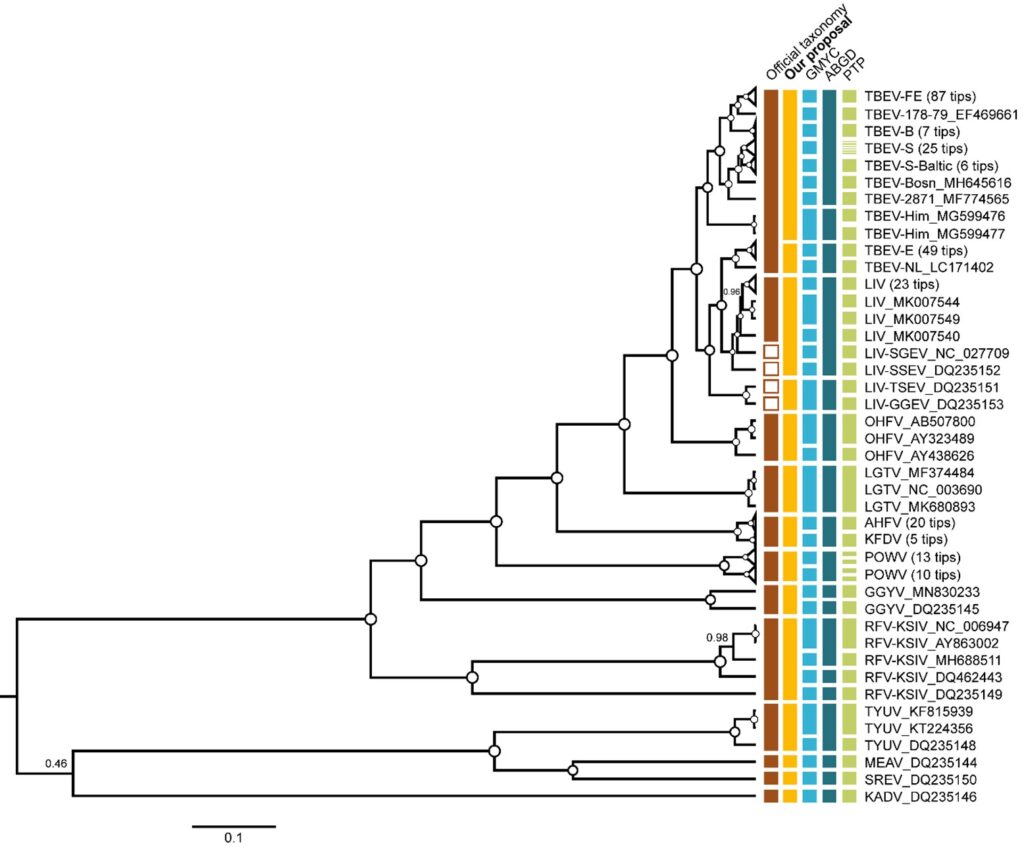
Figure 2: The phylogenetic tree of the TBFVs. The tree was reconstructed in BEAST using complete amino acid sequences (n = 278) of the polyprotein (3414 aa). For clarity, some of the wide clades were collapsed. Vertical bars to the right of tree tips indicate official classification (brown), our taxonomy proposal (orange), and delimitation results. Internal nodes with pp = 1 are marked as white circles, otherwise support values are shown by numbers ranging from 0 to 1.
Comparison of the Other Virus Particularities
We reviewed the literature for the other virus species criteria: natural and experimental host range, cell and tissue tropism, pathogenicity, vector specificity, and antigenicity. We have put all data on TBEV and LIV particularities observed into a table:
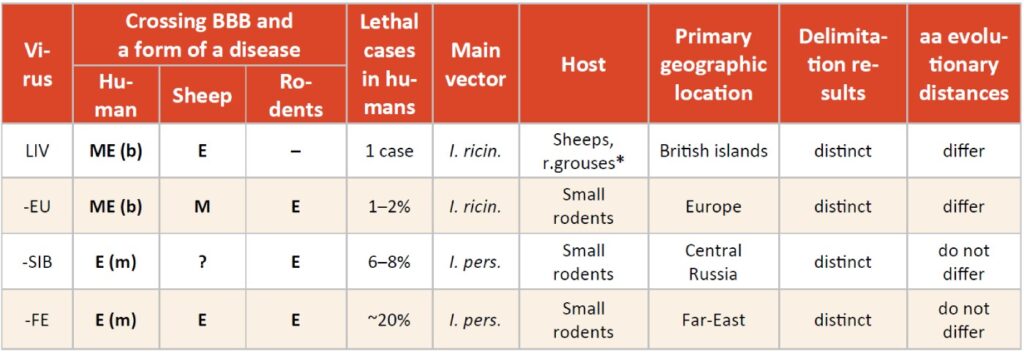
Table 1: Disease forms: E – encephalitis, M – meningitis, ME – meningoencephalitis; (m – monophasic, b – biphasic). *Rodents produce very low levels of viraemia as a result of LIV infection and do not support viraemic transmission as well.
As we can see, in some particularities, TBEV-EU is different from other TBEV subtypes and LIV, and sometimes it was closer whether to LIV or other TBEV members. We highlight two striking differences: reservoir host and pathogenicity in sheep. Unlike all TBEV subtypes, LIV is primarily found in red grouses and sheep inducing encephalitis and high mortality rate in both (78% in red grouses,4 5–60% in sheep,7 not small rodents. Although rodents such as field voles (Microtus agrestis), bank voles (M. glareolus) and wood mice (Apodemus sylvaticus) raised an antibody response to infection, they could not produce a substantial viremia and did not support non-viremic transmission between co-feeding ticks.5 This leads to the fact that LIV has patchy spatial distribution with different combinations of reservoir hosts occurring. This is exactly the opposite of the TBEV transmission patterns and natural foci structure formed by primarily small rodents. Concerning pathogenicity, in experiments with sheep, Votiakov et al9 demonstrated that TBEV-EU did not cross a blood-brain barrier (BBB) without lethal cases contrasting TBEV-FE.
Our Proposal
We believe that the differences described above are sufficient to delineate TBEV-E and LIV (+ SSEV and SGEV) from the joint TBEV clade into two distinct species. For the rest of the TBEV subtypes (TBEV-FE, -SIB, -BKL, HIM), we proposed to classify as a single species. TSEV and GGEV can be combined into a single species taxon. After the first round of the revision, we are preparing new species names in a binomial format different from those proposed in a preprint.1
Literature
- Bondaryuk AN, Andaev EI, Dzhioev YP, Zlobin VI, Tkachev SE, Bukin YS. Delimitation of the Tick-Borne Flaviviruses. Resolving the Tick-Borne Encephalitis and Louping-Ill Virus Paraphyletic Taxa. bioRxiv. 2021.2006.2011.448023. doi:10.1101/2021.06.11.448023. Published in February 2022: Mol. Phylogenet. Evolution 2022, 169, 107411, doi/org.10.1016/j.ympev.2022.107411
- Charrel RN, Zaki AM, Attoui H, et al. Complete coding sequence of the Alkhurma virus, a tick-borne flavivirus causing severe hemorrhagic fever in humans in Saudi Arabia. Biochem Biophys Res Commun. 2001;287(2):455-461. doi:10.1006/bbrc.2001.5610
- Fujisawa T, Barraclough TG. Delimiting species using single-locus data and the Generalized Mixed Yule Coalescent approach: a revised method and evaluation on simulated data sets. Syst Biol. 2013;62(5):707-724. doi:10.1093/sysbio/syt033
- Gilbert L. Louping ill virus in the UK: a review of the hosts, transmission and ecological consequences of control. Exp Appl Acarol. 2016;68(3):363-374. doi:10.1007/s10493-015-9952-x
- Gilbert L, Jones LD, Hudson PJ, Gould EA, Reid HW. Role of small mammals in the persistence of Louping-ill virus: field survey and tick co-feeding studies. Med Vet Entomol. 2000;14(3):277-282. doi:10.1046/j.1365-2915.2000.00236.x
- Grard G, Moureau G, Charrel RN, et al. Genetic characterization of tick-borne flaviviruses: new insights into evolution, pathogenetic determinants and taxonomy. Virology. 2007;361(1):80-92. doi:10.1016/j.virol.2006.09.015
- Jeffries CL, Mansfield KL, Phipps LP, et al. Louping ill virus: an endemic tick-borne disease of Great Britain. J Gen Virol. 2014;95(Pt 5):1005-1014. doi:10.1099/vir.0.062356-0
- Tang CQ, Leasi F, Obertegger U, Kieneke A, Barraclough TG, Fontaneto D, 2012. The widely used small subunit 18S rDNA molecule greatly underestimates true diversity in biodiversity surveys of the meiofauna. PNAS. 2012;109(40):16208-16212 doi:10.1073/pnas.1209160109
- Votiakov VI, Zlobin VI, Mishaeva NP. Tick-borne encephalitis of Eurasia (ecology, molecular epidemiology, nosology, evolution). Nauka, Novosibirsk; 2002.
Author: Dr. Artem N. Bondaryuk
Limnological Institute SB RAS, Ulan-Batorskaya 3, 664033, Irkutsk, Russia;
Irkutsk Antiplague Research Institute of Siberia and the Far East, Trilisser 78, 664047, Irkutsk, Russia.
Editor: Dr. Michael Bröker
Compiled: December 2021