Background
Tick-borne encephalitis virus can cause an inflammation of the central nervous system. While there are
currently no specific treatments, inactivated virus vaccines containing aluminum hydroxide as adjuvant are available and effectively prevent infection also from heterologous TBEV strains.
The development of adaptive responses evoked by the TBE vaccine has been investigated in detail; Aberle et al. showed induction of envelope protein-directed antibodies and of TBEV-specific CD4+ T cells in vaccinated individuals.1 However, a full picture of the innate immune responses induced by the vaccine is yet to be presented. The successful induction of innate immune responses has repeatedly been shown to correlate with the level of protection conferred by several vaccines.2,3 As the TBE vaccine has been proven to have a high field effectiveness, the investigation of the specific pathways it induces could help delineate vaccine-induced innate immune functions associated with a favorable vaccination outcome.
In this study, we aimed to delineate pattern recognition receptors (PRRs) and immune signaling pathways engaged by the TBE vaccine, in particular by inactivated TBEV (I-TBEV), the main component of the vaccine. An in vitro system consisting of human peripheral blood mononuclear cells (PBMCs), developed to assess the conformity of different TBE vaccine batches, was employed.4 The human PBMCs were exposed to I-TBEV or live TBEV and the cellular responses assessed by transcriptomic analysis.
Results
The results showed that both live and inactivated TBEV induced a distinct interferon-dominated signature in PBMCs. The stimulants induced partially overlapping expression signatures (Figure 1A): differentially expressed genes (DEGs) showing similar regulation in both treatment groups accounted for 71.5% of the total, while 28.5% of the genes showed opposite transcriptional signatures in I-TBEV- and live virus-treated cells. Hierarchical clustering identified 5 major clusters (Figure 1B). 3 clusters included genes similarly upregulated (clusters c and e: pathways related to the antiviral and IFN response and RIG-I-like receptor (RLR) signaling) or downregulated (cluster b: functions involved with RNA metabolism) in both treatment groups. 2 clusters included DEGs downregulated in I-TBEV-treated cells and upregulated in live TBEV-treated cells (cluster a: functions related to lipid metabolism), or vice versa (cluster d: functions involved in the immune response).
After having established the distinctive transcriptional profile of (I-)TBEV-stimulated cells, we sought to determine which pattern recognition receptors, once triggered, led to the identified responses. Using inhibitors of downstream factors of PRRs, we demonstrated the involvement of RLRs in I-TBEV sensing in cryopreserved PBMCs. Inhibition of MyD88 – an adapter protein downstream of Toll-like receptors – did not affect the expression of I-TBEV-induced genes, while inhibition of TBK1/IKKε – factors downstream of RLRs – halted the signaling cascade.
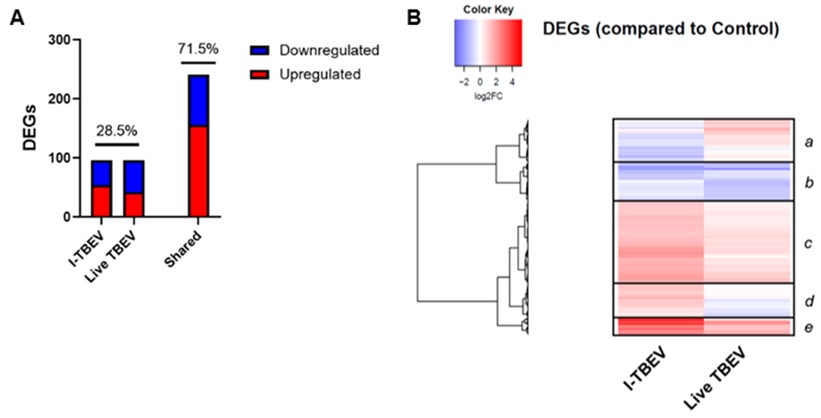
Figure 1: Transcriptional analysis of PBMCs stimulated with I-TBEV and live TBEV. After treatment with inactivated TBEV (I-TBEV) or live TBEV, the cells were lysed and processed for RNA sequencing. (A) Representation of differentially expressed genes (DEGs) with distinct or overlapping change in expression in the I-TBEV and live TBEV treatment groups as compared to the control. (B) Heatmap showing the fold change (FC) of DEGs in the two treatment groups, normalized to the control group (set as FC = 1) and hierarchically clustered.
DiscussionThe analysis of responses in human PBMCs induced by inactivated and live TBEV showed that the transcriptional profiles induced by the two treatments were mostly similar, with an interferon-dominated immune profile and the upregulation of selected inflammatory genes. Differences in responses arose for selected immune pathways (induced by the inactivated virus only) and for functions involved in lipid metabolism (upregulated only upon incubation with the live virus). These differences may be ascribed to the intrinsic nature of the replicating virus, which acts to suppress certain immune responses and to induce intracellular membrane rearrangements. Overall, given the induction of interferon responses by the inactivated virus at similar or higher levels than those induced by the live virus, I-TBEV stands out as a potent vaccine component.
The activation of RIG-I-like receptors, a predominant mechanism for cellular recognition of flaviviruses, was identified as the main trigger of the innate responses in our in vitro system for both live and inactivated TBEV. RLRs should not be triggered by the inactivated virus, since these receptors classically recognize nucleic acids produced during virus replication. However, activation of RIG-I by panhandle RNA (a partially circularized structure) was demonstrated for influenza A virus.5 Given the presence of cyclization elements in TBEV (and other flaviviruses) RNA, we hypothesize that, after the uncoating of the (inactivated) virus, such structures can be recognized by RIG-I in the cytosol.
The cellular responses to the final (adjuvanted) TBE vaccine formulation are yet to be unveiled: the alum adjuvant appeared not to be compatible with the viability of PBMCs. Further studies are required to confirm our findings in mouse models and in immune cells from vaccinated individuals.
Our study provides new insights into how I-TBEV activates the innate immune system in vitro. Understanding the mechanism of action through which a virus particle interacts with the immune system could (1) indicate whether critical viral components are retained during the manufacturing process of inactivated virus vaccines, (2) allow the selection or design of cellular platforms to be used for assessing the potential of the vaccine in vitro, and (3) provide leads for the design of effective vaccines for other pathogens.
Literature- Aberle JH, Schwaiger J, Aberle SW, et al. Human CD4+ T Helper Cell Responses after Tick-Borne Encephalitis Vaccination and Infection. PLoS One. 2015;10(10):e0140545. doi:10.1371/journal.pone.0140545
- Rechtien A, Richert L, Lorenzo H, et al. Systems Vaccinology Identifies an Early Innate Immune Signature as a Correlate of Antibody Responses to the Ebola Vaccine rVSV-ZEBOV. Cell Rep. 2017;20(9):2251-2261. doi:10.1016/j.celrep.2017.08.023
- Andersen-Nissen E, Fiore-Gartland A, Ballweber Fleming L, et al. Innate immune signatures to a partially-efficacious HIV vaccine predict correlates of HIV-1 infection risk. PLoS Pathog. 2021;17(3):e1009363. doi:10.1371/journal.ppat.1009363
- Signorazzi A, Etna MP, Coccia EM, Huckriede A. In vitro assessment of tick-borne encephalitis vaccine: Suitable human cell platforms and potential biomarkers. ALTEX. 2021;38(3):431-441. doi:10.14573/altex.2010081
- Lee MK, Kim HE, Park EB, et al. Structural features of influenza A virus panhandle RNA enabling the activation of RIG-I independently of 5′-triphosphate. Nucleic Acids Res. 2016;44(17):8407-8416. doi:10.1093/nar/gkw525
Author: Dr. Aurora Signorazzi
Dr. Aurora Signorazzi is a virologist currently working as viral safety scientist at Janssen Vaccines and Prevention, the Netherlands. The above research was conducted as part of her PhD thesis titled “In vitro approaches for the evaluation of human vaccines”, performed at the laboratory of Virology and Immunology of the Rijksuniversiteit Groningen under the supervision of Prof. Dr. Anke Huckriede, within the EU-funded consortium Vac2Vac.
Compiled: September 2021