Franz-Xaver Heinz and Karin Stiasny
Key Points:
- TBEV-particles are assembled in an immature, noninfectious form in the endoplasmic reticulum by the envelopment of the viral core (containing the viral RNA) by a lipid membrane associated with two viral proteins, prM and E.
- Immature particles are transported through the cellular exocytic pathway and conformational changes induced by acidic pH in the trans-Golgi network allow the proteolytic cleavage of prM by furin, a cellular protease, resulting in the release of mature and infectious TBE-virions.
- The E protein controls cell entry by mediating attachment to as yet ill-defined receptors as well as by low-pH-triggered fusion of the viral and endosomal membrane after uptake by receptor-mediated endocytosis.
- Because of its key functions in cell entry, the E protein is the primary target of virus neutralizing antibodies, which inhibit these functions by different mechanisms.
- Although all flavivirus E proteins have a similar overall structure, divergence at the amino acid sequence level is up to 60 percent (e.g. between TBE and dengue viruses), and therefore cross-neutralization as well as (some degree of) cross-protection are limited to relatively closely related flaviviruses, such as those constituting the tick-borne encephalitis serocomplex.
Introduction
Tick-borne encephalitis virus (TBEV) has played a pioneering role in the history of flavivirus structural biology, being the first of these viruses for which a high atomic resolution structure of the envelope glycoprotein E was determined1 (Figure 1A). This undertaking started 1987 in a collaboration between researchers at the Institute of Virology, University of Vienna, (now Center for Virology, Medical University of Vienna), and the Department of Biochemistry, Harvard University. The work took several years and required the purification of a total of 49 mg of a soluble form of the TBEV E protein (sE) that was isolated by trypsin cleavage from 401 mg of purified infectious TBEV. To obtain these amounts, 34,300 embryonated eggs were used for the preparation of primary chick embryo cells that were required for growing the virus. First structural details became visible in 1993, and the complete study was finally published in 1995.1
The structure of sE was a great surprise because of its unexpected features. In stark contrast to the prototypic influenza envelope glycoprotein (hemagglutinin, HA), which forms spiky projections of HA trimers at the viral surface, the TBEV E protein is an antiparallel dimer that is oriented horizontally to the viral membrane (Figure 1 A,B). Each of the monomeric sE subunits contains three domains (DI, DII, and DIII) that are connected to each other and the membrane-associated part of the protein by flexible linker regions. It took eight additional years until another flavivirus E protein structure (the dengue virus E protein) was published.2 Meanwhile, atomic resolution structures of E proteins are available for several of the most important human pathogenic flaviviruses, including dengue viruses, West Nile virus (WNV), Japanese encephalitis virus (JEV), Zika virus and Yellow fever virus (YFV),2-11 which gives the name to the genus Flavivirus in the family Flaviviridae.12 All of these structures have the same overall protein architecture as the TBEV E protein.
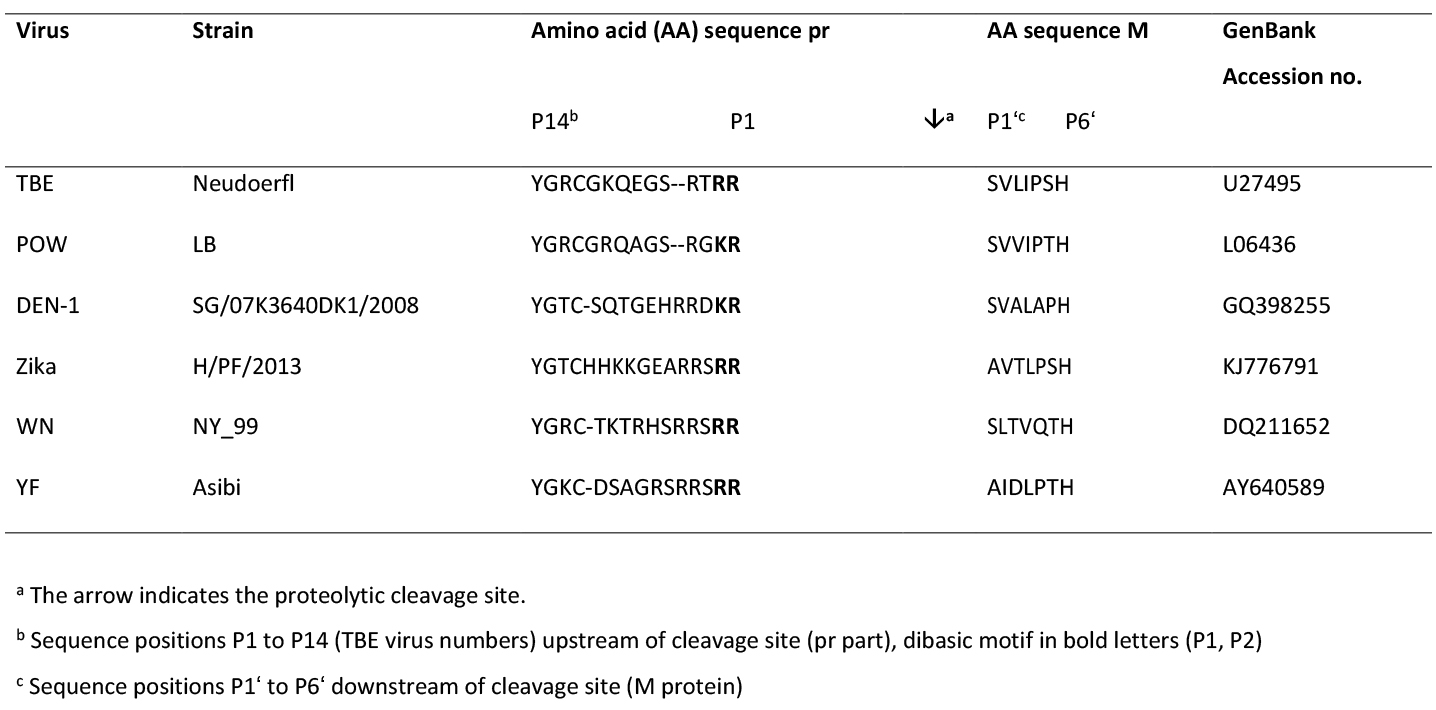
Figure 1: Structural organization of flaviviruses
Click the image above to enlarge
(A,B) Ribbon diagrams of the TBEV sE dimer [PDB code: 1SVB, (1)] and full-length E dimer [PDB code: 5O6A,26]. (A) Top view. (B) Side view. Color code E: domain I (DI), red; domain II (DII), yellow; domain III (DIII), blue; fusion loop (FL), orange; stem, green; membrane anchor, grey.
(C,D) Schematic representations of immature (C) and mature (D) virus particles.
(E,F) Electron cryo-microscopy structures of dengue virus serotype 1 particles. (E) Immature virion [PDB code: 4B03, 21]. (F) Mature virion [PDB code: 4CCT, (21)]. The prM proteins are shown in purple, and the E proteins in gray. (G) Electron cryo-microscopy structure of TBEV [PDB code: 5O6A,26], with individual domains of E colored as in A to D. One raft consisting of 3 parallel E dimers is encircled in white.
Panels A, B, E and F were prepared with PyMOL (Schrödinger LLC), panel G with UCSF Chimera [119, http://www.rbvi.ucsf.edu/chimera/].
In terms of their structure, flaviviruses are today among the best-studied enveloped viruses. Importantly, new technologies and instrumentation have led to the elucidation of structural details not only of the isolated E protein but also of whole virus particles using electron cryomicroscopy (cryo EM). Structures of both immature and mature virions are available for closely related mosquito-borne flaviviruses (such as dengue, West Nile, Japanese encephalitis, and Zika viruses)13-25 and form the basis for understanding the viral life cycles and interactions with antibodies at a molecular level. Recently, a high-resolution cryo-EM structure of mature TBEV was published by Fuzik et al.,26 providing for the first time details of the particle organization and interactions of proteins in a flavivirus transmitted by ticks (Figure 1B,G).
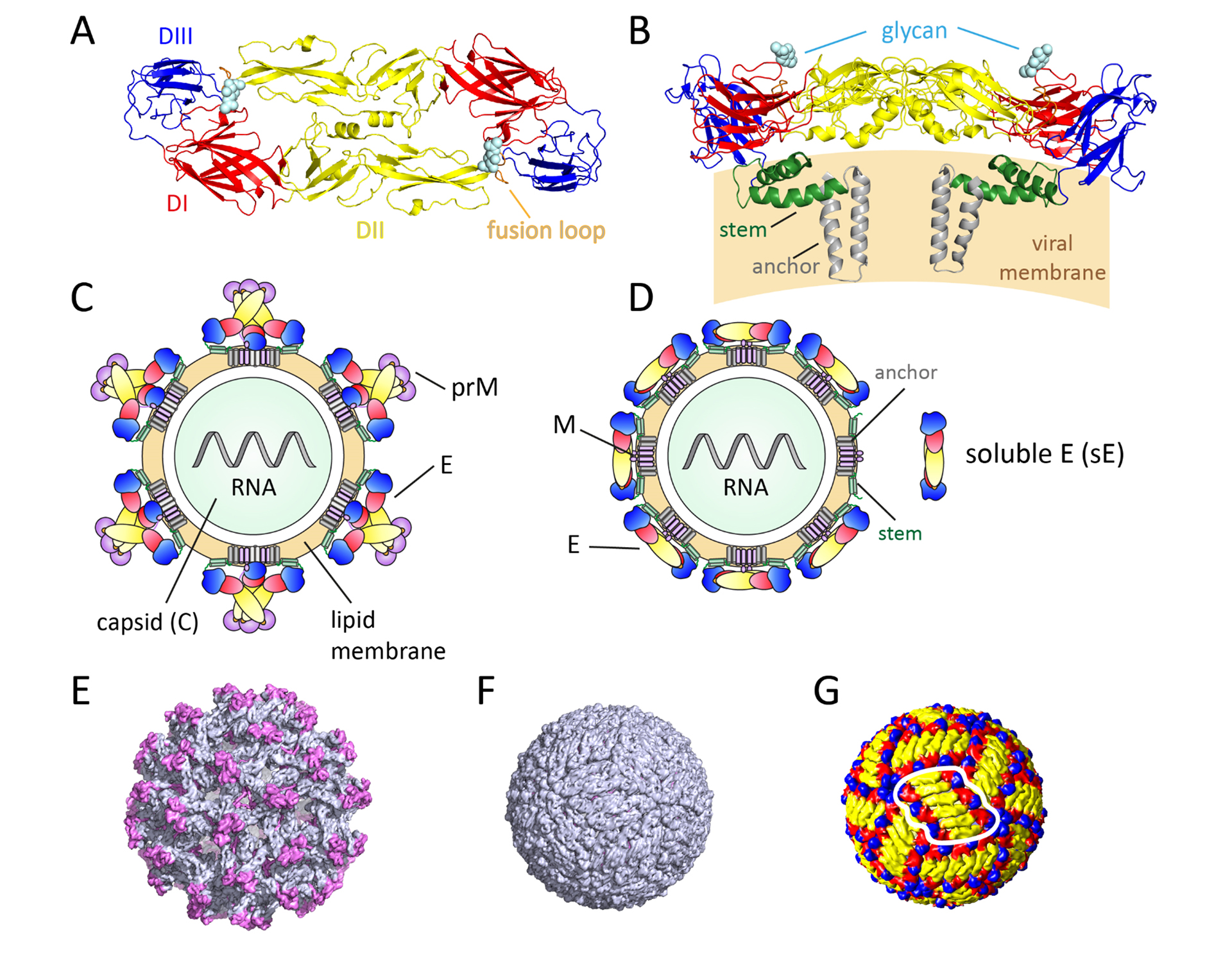
Figure 2: Life-cycle of flaviviruses
Click the image above to enlarge
Left: Virus entry. Viruses are taken up by receptor-mediated endocytosis and low pH in endosomes triggers viral membrane fusion, resulting in the release of the viral genome into the cytoplasm. Protein translation and RNA replication occur at virus-induced ER membranes.
Right: Virus assembly, maturation and release. Formation of immature virions takes place by a budding process into the ER. As a byproduct, subviral particles are formed that are devoid of a nucleocapsid. Particles are transported through the exocytic pathway. The acidic pH in the TGN causes a major structural rearrangement that leads to the formation of an E herringbone-like arrangement that is characteristic of mature virions (see Figure 1) and exposes the furin cleavage site in prM. The cleaved-off pr segment of prM remains associated with E at acidic pH but falls off at the neutral pH of the extracellular fluid upon secretion of the particles.
Color code of prM and E as in Figure 1.
Here, we review the structure of TBEV with a focus on the role of E in the viral life cycle and as a major determinant for the induction of virus-neutralizing antibodies. These properties are discussed in the context of what is known for other flaviviruses, in order to provide a more rounded picture of TBEV structure-function relationships and to emphasize the gaps that still exist in our understanding of the structural foundations of TBEV biology.
Virus particle structures and life cycle
Virus assembly, maturation and release
Virus assembly takes place in the endoplasmic reticulum (ER) and leads to the formation of immature particles (Figure 1C,E and Figure 2).27 This first assembly product contains three structural proteins: C (capsid), forming an ill-defined spherical core together with the viral genomic RNA, and two membrane associated proteins, prM (precursor of M) and E in a heterodimeric complex. Trimers of these heterodimers form spikes at the surface of immature particles that are non-infectious (Figure 1C,E).
Studies with TBEV have provided evidence that prM functions as a chaperone for the correct folding of E during its biosynthesis, at least in certain cellular environments.28 Experiments with recombinantly expressed prM and E proteins in mammalian cells (COS-1) revealed that heterodimerization of the two proteins occurs rapidly and is important for the final folding steps. On the one hand, E apparently requires prM to reach its native conformation efficiently and on the other hand, prM needs E for rapid signal sequence cleavage at its N-terminus during viral polyprotein processing. After their formation in the ER, immature virus particles are transported through the exocytic pathway of the cell. As a crucial step of virus maturation, the prM protein is cleaved in the trans-Golgi network (TGN) by the cellular protease furin, generating membrane-anchored protein M and the proteolytic fragment pr.
Increasing the pH in the TGN of TBEV-infected cells by acidotropic agents (such as ammonium-chloride) or by bafilomycin A1 (a specific inhibitor of the vacuolar type H+ ATPase) led to the release of immature particles with a 20 – 50 fold lower specific infectivity and hemagglutination (HA) activity than mature viruses.29 This suggested that a conformational change in the prM-E complex is induced by the slightly acidic pH in the TGN, which is required for furin cleavage. Evidence that the maturation cleavage is conferred by the TGN-resident protease furin was obtained in experiments with a furin-deficient human cell line (LoVo), which produced only immature viruses, as well as a specific furin inhibitor that blocked furin cleavage, and by in vitro cleavage experiments with recombinant furin.30 Treatment of immature TBEV particles with furin resulted in a 100-fold increase in specific infectivity and the acquisition of hemagglutination as well as membrane fusion activities.
Importantly, furin cleavage itself did not require an acidic pH, but the conformational change exposing the cleavage site in the prM-E complex was acid pH-dependent. The low-pH-induced reorganization of the protein complex was shown to be irreversible in the case of TBEV30 but appears to be reversible in the case of dengue viruses.31
The furin cleavage site of TBEV corresponds to a consensus sequence also found in other flavivirus prM proteins (Table 1). The dependence of virus maturation on this conserved sequence element in prM was demonstrated directly by a genetic approach. A specific mutation in the furin recognition sequence engineered into an infectious TBEV clone (resulting in the deletion of one of the arginines at P1,P2; Table 1) did not impair the assembly of immature particles but completely abolished infectivity.32 Infectivity could be restored by in vitro trypsin cleavage, which is likely to cleave at one of the R residues that was retained at the furin cleavage site (Table 1).
So far, the structure of immature virions has only been determined for mosquito-borne flaviviruses, which were shown to carry 60 spikes of trimers of prM-E heterodimers. 16,19,21,31 Considering the high degree of structural conservation of viral proteins and mature particles, it is justified to assume that immature TBE virions are similar to those of mosquito-borne flaviviruses. In the course of exocytosis of immature viral particles, the acidic pH in the TGN causes a major re-arrangement of the viral glycoprotein interactions, resulting in the conversion of the trimeric prM-E spikes into a herringbone-like shell of 90 E protein dimers (Figure 1G). Data obtained with dengue virus show that the pr fragment remains associated with the particles at acidic pH after furin cleavage but dissociates at neutral pH when the particles are released from the cells (Figure 2).31
Mature virions display the herringbone-like arrangement of E that was induced in immature particles when encountering the low pH in the TGN. The release of the pr fragment leaves E in a metastable conformation, poised to undergo dramatic low pH-induced structural changes that mediate viral fusion in endosomes upon virus entry (see below). The function of prM and the pr fragment is thus to protect E in the acidic TGN and to avoid membrane fusion already at this stage of the viral life cycle.33
The static pictures of fully immature and fully mature particle structures determined by cryo EM cannot be reconciled with all experimental data obtained in studies of flavivirus entry and virus interactions with antibodies.34,35 First, some antibodies binding to seemingly inaccessible (cryptic) epitopes in E neutralized viral infectivity in various flavivirus systems. These observations led to the concept of ‘virus breathing’ as a consequence of envelope glycoprotein dynamics36, reflecting the metastable nature of E which transiently exposes otherwise buried protein surfaces within the E dimer or at the inter-dimer contact regions in the virion. Retrospectively, antibody-induced conformational changes, described for TBEV already in 1984, are also likely due to E protein dynamics and virus breathing.37 Secondly, many data indicate that virus particles released from infected cells are a heterogeneous mixture of immature, partially mature and fully mature particles.38,39 As a specific structural feature, partially mature and breathing particles expose the viral membrane, which has been shown to be a target for interactions with cellular lipid receptors that can mediate cell entry.40,41
It can be hypothesized that an ensemble of heterogeneous particles in combination with virus breathing may be important for flaviviruses to infect different tissues in their invertebrate and vertebrate hosts.34 Heterogeneity may also be required to maintain these viruses in their natural cycles and constitute a powerful means to adapt to new environments or to acquire new pathogenic properties, such as those observed in the recent Zika virus epidemic.42,43
Subviral particles
Flavivirus-infected cells do not only secrete complete virus particles but also subviral particles that are non-infectious and smaller than whole viruses but have similar HA activity. Because of these properties they were described as ‘slowly sedimenting hemagglutinin’ (SHA) in the flavivirus literature.44,45
Noninfectious subviral particles of TBEV were produced in recombinant form by the co-expression of the two viral glycoproteins prM and E in COS-1 cells.46,47These particles [designated ‘recombinant subviral particles’ (RSPs)] were secreted from transfected cells and had a density of approximately 1.14 g/cm³.48,49 They were sensitive to disintegration by the detergent Triton X 100, consistent with the presence of a lipid membrane carrying the two viral envelope proteins.48 The formation of RSPs could also be achieved by the expression of prM and E from separate plasmids, but was not possible with a soluble form of E that lacked its membrane anchor.47 More detailed mapping studies allowed the identification of the so-called stem together with the first trans-membrane regions of E to be essential for particle formation.50
The ER was shown to be the site of assembly of RSPs by biochemical and electron microscopical analyses.51 In addition to the rough ER, RSPs were observed in the smooth ER and down-stream compartments of the secretory pathway. Approximately 75% of the particles had a diameter of 30 nm, but a number of larger particles and tubular structures were also seen in vesicular compartments of transfected cells.51
It is an important conclusion of these studies that the formation of prM-E heterodimers and their lateral interactions are sufficient to drive the budding of membrane-containing virus-like particles at the ER membrane, in the absence of any interactions with viral RNA or a capsid.
The 30-nm RSPs were the first flaviviral particles for which a cryo-EM structure was determined.52 The 19Å resolution map revealed an arrangement of E protein dimers in a T=1 icosahedral surface lattice (different from that of the virion, Figure 1G) and allowed the definition of interaction sites between E dimers, positions of M relative to E, and the assignment of transmembrane regions of E and M. When the prM furin cleavage site was deleted in the plasmid construct for RSP production by mutagenesis, a substantial number of particles were observed that had the same size as whole immature virions (diameter 60 nm), in addition to the 30-nm particles described before.53 It was therefore concluded that the primary assembly products in prM-E expressing cells are immature particles of both size classes, but in their mature forms (i.e. after prM cleavage) the larger particles are less stable and therefore seen as a minority compared to the 30-nm particles secreted from transfected cells. Apparently, alternative assembly products can be formed by prM-E interactions. The role of subviral particles in natural TBEV infections of ticks and/or mammalian hosts remains to be elucidated.
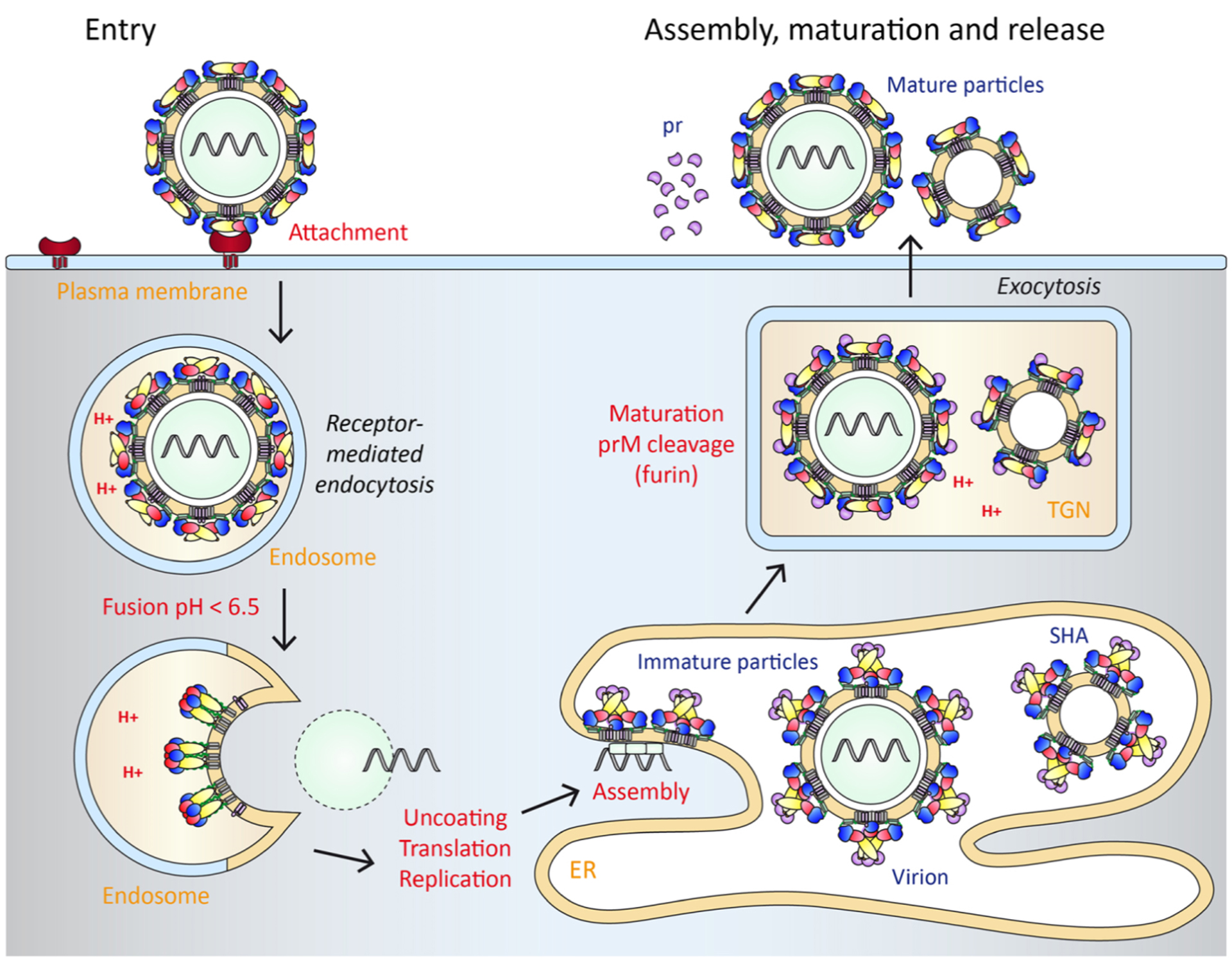
Figure 3: Post-fusion structure of E and fusion mechanism
Click the image above to enlarge
(A) Ribbon diagrams of E monomers in their pre- and post-fusion conformations, revealing the relocation of domain III (indicated by a blue arrow in the pre-fusion conformation).
(B) Ribbon diagram of the trimeric post-fusion structure of TBEV sE [PDB code: 1URZ,87].
(C) Schematic of steps involved in flavivirus membrane fusion. Panel I: Metastable E dimers at the viral surface. Panel II: Low-pH-induced dimer dissociation, exposure of the FL and interaction with the endosomal membrane. Panel III: Relocation of domain III and trimer formation. Panel IV: Stem zippering and hemifusion intermediate. Panel V: Final post-fusion structure of E and opening of a fusion pore.
Color code of E as in Figure 1.
Ribbon diagrams were prepared with PyMOL (Schrödinger LLC).
The E protein in RSPs appears to be structurally and functionally identical to that at the surface of whole TBE virions.48 As a consequence, RSPs proved to be a valuable non-infectious model system to assess biological properties of E, including membrane fusion and antigenic structure (see below: Structure and functions of E – Virus entry and membrane fusion; Antigenic structure of TBEV and virus neutralization).54-60 Importantly, their particulate nature also makes them an excellent candidate for use as a recombinant vaccine antigen, as shown by mouse immunization and challenge experiments.61 In these experiments, the immunogenicity of RSPs was compared with soluble E dimers, E rosettes formed by detergent removal after solubilization of the viral membrane, and whole formalin-inactivated purified TBEV. With respect to both the extent of antibody induction and protection from challenge, the RSPs were equivalent to the inactivated virus vaccine. This high immunogenicity is most likely due to the presentation of multiple copies of the native E protein on a large particulate carrier, mimicking its presentation on whole virus particles. Similar conclusions were also derived from a DNA immunization study in mice.62 Plasmid constructs giving rise to secreted RSPs were superior to those expressing a secreted C-terminally truncated E dimer, or a non-secreted full-length form of E.
Figure 4: Binding sites of TBEV E-specific mAbs
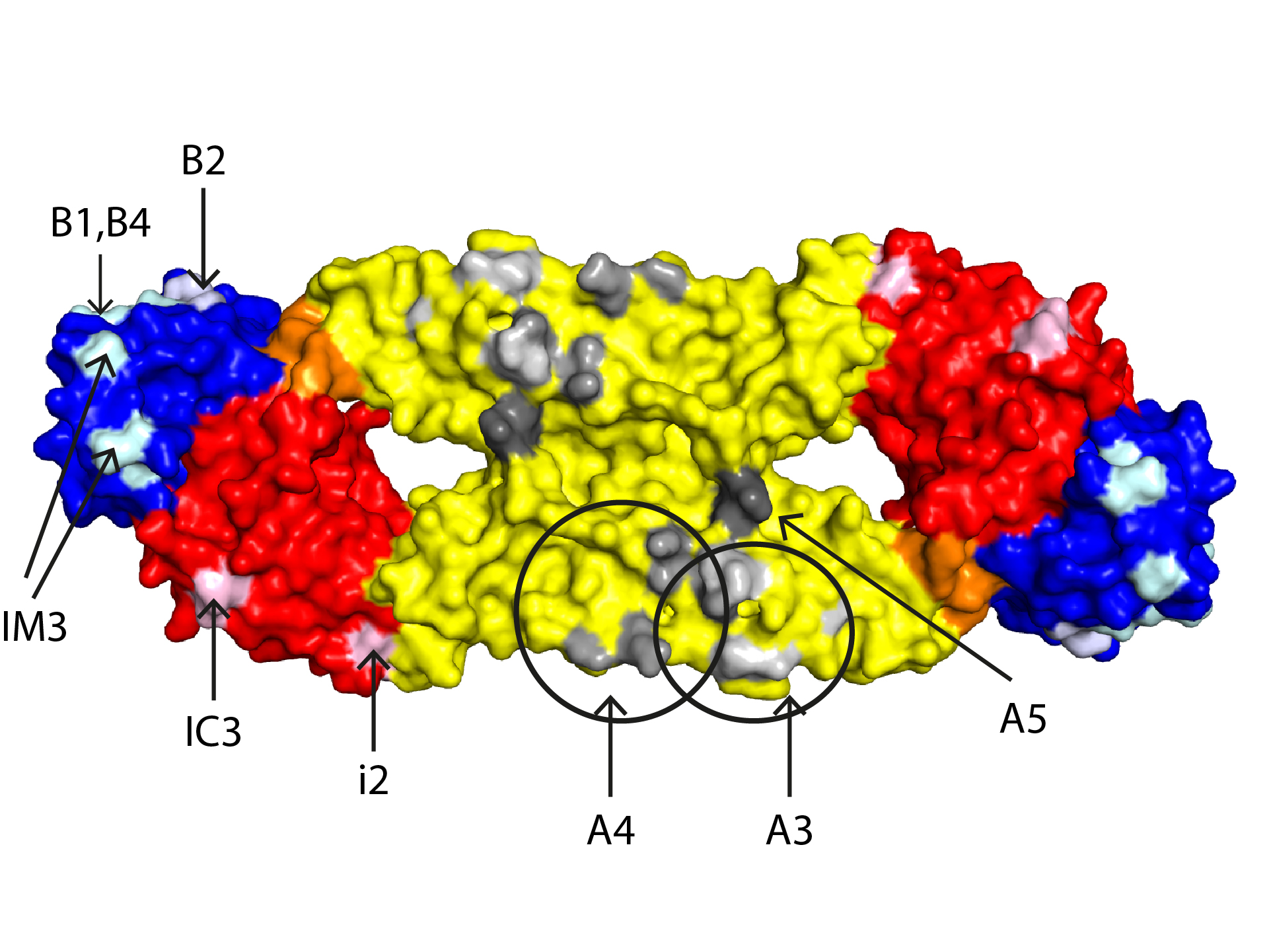
Click the image above to enlarge
Surface representation of the TBEV sE dimer [PDB code: 1SVB,1] with the location of amino acids involved in binding sites of neutralizing mAbs. Epitopes are labeled only on one of the two monomers and the mAbs are designated according to references.92,95
Color code of E as in Figure 1.
The figure was prepared with PyMOL (Schrödinger LLC).
Figure 5: Antigenic relationships of flaviviruses
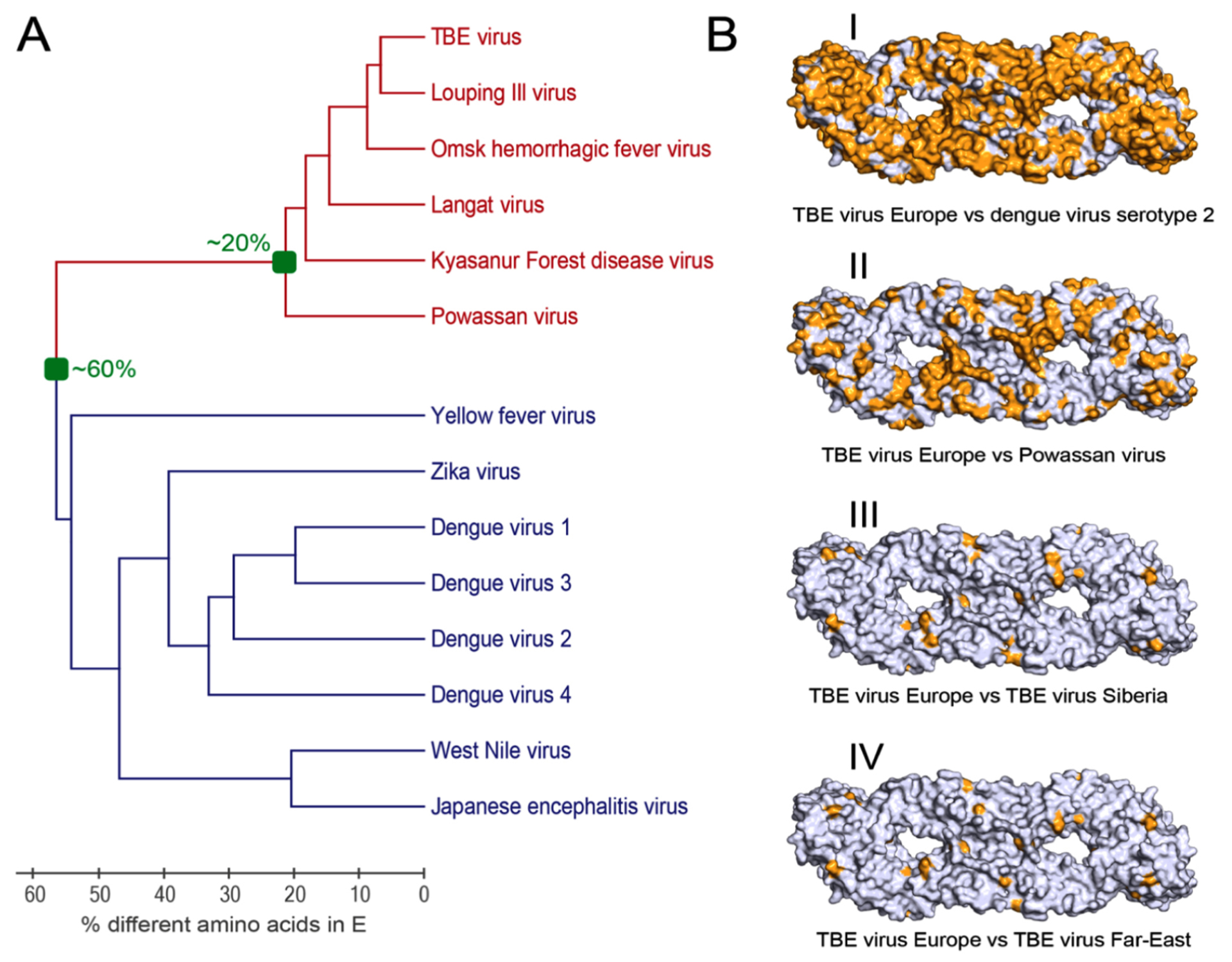
Click the image above to enlarge
Dendrogram based on amino acid differences of the TBEV serocomplex (red) and other flaviviruses (blue) (MAFFT Alignment: http://www.ebi.ac.uk/Tools/msa/mafft/).
TBEV (GenBank accession no. U27495), Louping Ill virus (NC_001809), Powassan virus (L06436), Omsk hemorrhagic fever virus (NC_005062), Langat virus (AF253419), Kyasanur Forest disease virus (AY323490), yellow fever virus (AY640589), Zika virus (KJ776791), dengue virus serotype 1 (GQ398255),dengue virus serotype 2 (NC_001474), dengue virus serotype 3 (EU081190),dengue virus serotype 4 (GQ398256), West Nile virus (DQ211652), Japanese encephalitis virus (D90194)
Surface representations of the TBEV sE dimer (strain Neudoerfl, GenBank accession no. U27495, European subtype; PDB code: 1SVB, Rey et al., 1995) in pairwise comparisons with E of other viruses, displaying divergent surface-exposed amino acids in orange. Panel I: dengue virus serotype 2 strain 16681 (NC_001474). Panel II: Powassan virus strain LB (L06436). Panel III: TBEV strain Vasilchenko, Siberian subtype (AF069066). Panel IV: TBEV strain Sofjin, Far Eastern subtype (AB062064).
Panel B was prepared with PyMOL (Schrödinger LLC).
Structure and functions of E
The TBEV E protein has at least two essential functions in the viral life cycle (Figure 2), consistent with its prominent presentation at the viral surface. It is responsible for interactions with attachment factors and/or entry receptors at the plasma membrane of target cells, and it mediates viral membrane fusion after cellular uptake by receptor-mediated endocytosis. While TBEV membrane fusion has been studied in great detail, the search for viral receptors is still quite elusive, reminiscent of the situation described for flaviviruses in general.40
Cell attachment and receptors
Several sets of experiments have provided evidence that TBEV can use negatively-charged glycosaminoglycans (GAGs) such as heparan sulfate (HS) as an attachment factor in certain cells.53,63 Passaging of a virus isolate from ticks in BHK-21 cells resulted in the accumulation of mutations that were distributed over a large part of the upper and lateral surface of E including each of the three domains63 (Figure 1). Importantly, these mutations resulted in an increase of positive charges at the viral surface, increasing its affinity for BHK-21 cells. Growth of the mutant viruses, but not the wild type, could be inhibited competitively by heparin, confirming their adaptation and dependence on GAG-binding for entry. The increased affinity for GAGs was associated with a decrease in virulence in a mouse model and may be a general principle for attenuating flaviviruses. A connection between an increased binding to GAGs and attenuation was also observed for viruses of the JEV serocomplex64-66 and the 17D strain of the live yellow fever vaccine.67
The role of GAGs in TBEV entry was investigated in greater detail using mutant CHO cells that are deficient in the synthesis of GAGs.68 Interestingly, while virus binding to these mutant cells was much lower than that to normal CHO cells, no difference was observed in terms of cell infection. It was therefore suggested that HS is not required for the infection of CHO cells, and that one or more other receptors are required for virus entry into these cells.
Since the structure determination of E, the immunoglobulin-like domain III (Figure 1) has been hypothesized to be a site of receptor interactions not only for TBEV but for flaviviruses in general.1,69 This was primarily based on the fact that a number of mutations affecting flavivirus virulence were concentrated in this domain and that the so-called FG loop is enlarged to contain an RGD sequence in some mosquito-borne flaviviruses70/sup><, which is a characteristic ligand-binding motif for members of the integrin family of cell surface receptors.71 Experiments with recombinant domain III of Langat virus (a close relative of TBEV) have revealed that its addition to cells before infection resulted in a somewhat decreased virus growth, which was interpreted as evidence that DIII is involved in receptor binding.72 So far, however, there is no information as to possible interaction partners of DIII at the cell surface and further efforts will be necessary to get a more complete picture of TBEV receptor interactions.
In general, it is believed that flaviviruses may use different receptors in different tissues of the various invertebrate and vertebrate host species involved in natural transmission. It was recently shown for several flaviviruses (but not yet for TBEV) that not only the E protein but also lipids of the viral membrane may bind to cellular lipid receptors.73,74 They normally recognize apoptotic cells and control their removal by phagocytes.75,76 Hijacking these receptors by viruses to gain access to cells has therefore been designated apoptotic mimicry.41
Virus entry and membrane fusion
The presence of acidotropic agents such as NH4Cl or bafilomycin A1 early in infection had a strong inhibitory effect on the replication of TBEV, consistent with virus uptake by receptor-mediated endocytosis and the importance of an acidic endosomal compartment for viral membrane fusion.29 The acid pH-dependence of TBEV fusion activity was first demonstrated by Guirakhoo et al. 199177 and further studied in great detail using a combination of biochemical, structural, mutational, and functional studies.54,57,59,60,78-83 Chemical cross-linking experiments and sedimentation analyses demonstrated that the exposure to acidic pH caused a quantitative oligomeric rearrangement of metastable E dimers into stable trimers at the virion surface, with a pH threshold of 6.5,84 suggesting that this dimer-trimer transition provides the energy and drives the fusion of viral and endosomal membranes. Further biochemical studies85 indicated that the structural conversion of E was a two-step process, in which the acidic pH in endosomes first caused the dissociation of E dimers followed by an irreversible trimerization. Monoclonal antibody studies and mutational analyses provided evidence that the highly conserved sequence element in E, located at the tip of DII and now designated fusion loop (FL), was responsible for interacting with the endosomal target membrane as an initial step in membrane fusion.55,79
It was a key finding of these studies that the soluble E dimer (which dissociates into monomers at acidic pH) could be converted into a trimer in the presence of liposomes,79,86</a> laying the foundation for the crystallization of this post-fusion conformation and the determination of its atomic structure by X-ray crystallography87 (Figure 3). The structure revealed that the folding of the three domains is maintained but that their relative orientation is altered (Figure 3A,B). Specifically, DIII relocates from its position at the end of the dimer to the side of the trimer in such a way that a hairpin-like structure is formed (Figure 3A), in which the FL and the stem-anchor region of E would be juxtaposed in the full-length E trimer. This structure was reminiscent of the post-fusion structures of class 1 viral fusion proteins such as the influenza virus hemagglutinin and suggested – in combination with studies on fusion intermediates83/sup>< – that the TBEV fusion mechanism consists of several steps as depicted in Figure 3C. In this process, the acidic-pH-induced dissociation of E dimers leads to the exposure and interaction of the FL with the endosomal membrane, the relocation of DIII and the zippering of the ‘stem’ along DII in the trimer, thus driving the merger of the two membranes. Mutational analyses provided evidence for a specific molecular interaction at the N-terminal end of the stem and a pocket of DII which appears to be essential for the correct positioning of the stem for the zippering reaction.60
An important question in the context of TBEV fusion relates to the molecular switches that sense the acidic pH in endosomes and induce the fusogenic conformational change in E. Because of their pKa near the pH threshold of fusion, histidines have been hypothesized to play such a role in the fusion trigger. There are indeed five histidines (H146, 248, 287, 323, 438) that are absolutely conserved among flavivirus E proteins, suggesting an indispensable structural and/or functional role in the viral life cycles. The use of RSPs (see above) with mutated histidines at these positions allowed the identification of H323 as a key residue for triggering the acidic-pH-induced trimerization of E and concomitant membrane fusion.57 This residue is involved in intramolecular interactions at the interface between DI and DIII in the E dimer. Its protonation apparently facilitates E dimer dissociation and allows DIII to be released from its original position and to relocate as required for post-fusion trimer formation (Figure 3). Other conserved histidines were shown to be dispensable for fusion, but they may have critical roles in unrelated low-pH-driven processes of the viral life cycle, such as virus maturation (Figure 2).
Antigenic structure of TBEV and virus neutralization
Because of its functions in flavivirus attachment and entry as well as membrane fusion in endosomes (see above and Figure 2,3), the E protein is the major target and inducer of neutralizing antibodies, and all experimental data obtained with TBEV are consistent with this notion. Potently neutralizing and protective antibodies were induced by E solubilized from purified TBEV,61 confirming the primary role of such antibodies in the induction of a protective immunity.88 While soluble forms of E and even the isolated DIII were capable of inducing neutralizing antibodies,61,89 particulate or aggregated forms (E rosettes) had a much higher specific immunogenicity and would therefore be preferred vaccine antigens.61
Epitopes of TBEV protein E
More precise mapping of epitopes in E and their involvement in virus neutralization became possible with the preparation of TBEV-specific monoclonal antibodies (mAbs).90-92 By the application of these mAbs for immunochemical analyses and the selection of mAb-resistant virus mutants, topological and functional models of epitopes could be defined and epitopes involved in virus neutralization were structurally characterized.37,58,91-96 The elucidation of the crystal structure of the E dimer then allowed the precise localization of antibody-binding sites (Figure 4). It became apparent that neutralizing antibodies can be induced by each of the three domains, consistent with a plethora of publications on the antigenic structure of other flaviviruses [reviewed in references97-99].
The complexity of the antigenic structure cannot be understood completely on the basis of the isolated E protein. Studies with different soluble and particulate forms of E revealed a strong influence of its quaternary structure and specific display at the surface of virions.58 The TBEV data are fully consistent with those obtained by high-resolution structural analyses of antibody-E complexes of other flaviviruses [reviewed in references99,100]. Taken together, it can be concluded that TBEV, like all other flaviviruses, displays a continuum of antigenic sites at its surface with the potential of inducing neutralizing antibodies. Epitopes of such antibodies have been mapped to individual domains in E or were shown to be more complex and to comprise residues from adjacent domains, from both monomers in the dimer or even adjacent dimers in the herringbone arrangement of E at the viral surface (quaternary epitopes) [reviewed in references99-101].
Mechanisms of virus neutralization
The most apparent mechanism of virus neutralization by E-specific antibodies is the blocking of cell attachment, as shown for different flaviviruses.102 In addition, the inhibition of post-entry processes by antibodies bound to the internalized virus is likely to contribute to virus neutralization.102 This holds especially true for membrane fusion, which requires substantial relocations of protein domains (see above) that may be impeded by bound antibodies. Insights into such activities were provided by in vitro analyses of TBEV fusion inhibition by E-specific mAbs.82 Depending on the location of the bound antibody, early and late stages of the fusion process were impaired, by either blocking the initial interaction with the target membrane or by interfering with the required relocation of DIII and the formation of the post-fusion E trimer (see above). A special case are antibodies directed to the fusion loop at the tip of DII (Figure 1A) which – because of the high conservation of this structural element – are highly cross-reactive with E proteins from all flaviviruses.56 They react strongly with soluble forms of E and inhibit in vitro liposomal fusion,82 but they have virtually no neutralizing activity against TBEV. An explanation of this phenomenon is the fact that FL-specific antibodies are unable to react with intact mature TBE virions (‘cryptic epitopes’)56 and therefore cannot reach the endosomal compartment where fusion takes place. The cryptic nature of the FL may however differ among flaviviruses, depending on their stability and breathing behavior (see above). As a consequence, broadly flavivirus cross-reactive antibodies may display neutralizing activity against certain viruses only, a feature observed especially with dengue viruses.35,103
Antigenic relationships of TBEV with other flaviviruses
Even the most distantly related flaviviruses have approximately 40% identical amino acids in their E proteins (Figure 5A). Most of these residues, however, are located inside the protein whereas most of the surface-exposed and antigenically relevant residues differ among flaviviruses from different serocomplexes. This is visualized in a comparison of such residues in E of TBEV versus that of dengue virus serotype 2 (Figure 5B, panel I) which shows that almost the whole surface is different, explaining the lack of cross-neutralization between TBEV and flaviviruses of other serocomplexes (Figure 5A). The only patch of conservation includes the fusion loop, which is cryptic in TBEV and therefore inaccessible for antibodies (see above).
Cross-neutralization is, however observed within the TBEV serocomplex (Figure 5A), which also includes Louping Ill, Langat, Omsk hemorrhagic fever, Kyasanur Forest disease, and Powassan viruses. These viruses display a higher degree of conserved patches of amino acids at their surface that is responsible for cross-neutralization. Powassan virus is the most distant relative of TBEV in this serocomplex with approximately 20% sequence divergence in E (Figure 5A).
TBEV subtypes and strains
Comparison of virus strains from all areas of TBEV endemicity have revealed three major subtypes [European, Siberian, and Far Eastern101/sup><] which are sometimes also referred to as genotypes.104 Additional heterogeneity may exist and two further genetic lineages have been described.104,105 Overall, the amino acid sequence divergence observed in the E proteins of different TBEV subtypes does not exceed 6.9%.106 This is within the range of natural variation observed with other human-pathogenic flaviviruses (e.g. YFV 5%; WNV 7%). Pairwise comparisons of individual strains from different subtypes show that the differences are relatively small (Figure 5B). Variation observed within the subtypes is even smaller and does not exceed 1.8% for the European subtype.
The low degree of antigenic variation is an important aspect of vaccine usage. Experiments with serum samples obtained after vaccination with a European subtype TBE vaccine revealed no differences in the neutralization of European, Siberian or Far Eastern TBEV subtype strains, whereas neutralization of the closely related OHF virus (Figure 5A), was somewhat reduced.107 In a related study, a high degree of cross-protection between TBEV subtypes was also observed in mouse challenge experiments after immunization with vaccines based on European or Far-Eastern subtype strains.105,108 It was therefore concluded that a single vaccine will protect against all TBEV strains circulating in nature, similar to the situation with vaccines against other flaviviruses such as JEV and YFV.
Overall, the degree of cross-neutralization by polyclonal sera within the TBEV serocomplex (and other flavivirus serocomplexes) seems to follow the degree of amino acid conservation in E (Figure 5A). Observations made with some flaviviruses, however, indicate that differences at single amino acids can lead to substantial differences in virus neutralization, presumably due to influences of such mutations on virus envelope dynamics and the accessibility of certain epitopes.109,110 A similar variation, related to a single amino acid difference in E, was reported in a comparative study of vaccines that use different strains as seed viruses for vaccine production.111 Differences were found in the induction of antibodies that neutralize circulating strains of TBEV that could be related to a single amino acid difference (N52K) at the hinge region between DI and DII.
Fine specificities of antibody responses to TBEV
The mapping of epitopes in the E protein of TBEV and other flaviviruses with mAbs has provided us with deep insights into antigenic structure and details of antibody interactions with these viruses. In contrast, relatively little is known about the fine specificities of antibodies in polyclonal responses as well as their individual variation after TBEV infections and vaccinations. The issue was addressed by Jarmer et al.112 who deconstructed human antibody responses after TBEV infection and vaccination using immunoassays with recombinant proteins consisting of individual domains and domain combinations of E. Extensive variation was not only observed with respect to the extent of antibody formation but also with respect to the fine specificities of antibodies produced in the course of immune responses, suggesting that patterns of immunodominance are strongly influenced by individual-specific factors. Importantly, most of the neutralizing activity could be depleted from sera by the dimeric E protein, indicating that complex quaternary epitopes, depending on the herringbone-like arrangement of E dimers at the viral surface (Figure 1G), play only a minor role in the neutralizing antibody response, both after infection and vaccination.
In humans, it is currently unknown to what extent the fine specificities of antibody responses can be modulated by pre-existing antibodies (against homologous or heterologous flavivirus antigens) when present at the time of infection or vaccination. A mouse immunization study with the recombinant TBEV E protein dimer and passively administered monoclonal antibodies, however, revealed that such influences may be substantial.113 Mechanistically, the differences observed in antibody responses were related to shielding of epitopes in E by the co-administered mAb and to mAb-induced dissociation of the E dimer, resulting in the exposure of antigenic surfaces that would be cryptic in the native protein. It remains to be seen, whether human antibody responses may be modulated by similar mechanisms and whether the resulting changes in antibody fine specificity and composition can affect virus neutralization.
Perspectives and future research
The era of flavivirus structural biology was initiated by the X-ray structure determination of the TBEV E protein dimer1 and has now led to unprecedented insights into the organization and molecular changes of flavivirus particles in different phases of the viral life cycle.114-116 Although a high resolution particle structure of TBEV is now available in its mature form,26 the structure of immature particles has not yet been determined and will be a topic of future research. The same also holds true for investigations in the complex area of viral receptors. Recent data obtained with other flaviviruses suggest that populations of heterogeneous, partially mature but infectious virus particles may be produced in different hosts and tissues involved in the viral life cycles. Such heterogeneity in combination with the phenomenon of virus breathing is a mechanism that modulates the viral surface and thus increases potential interaction sites with cellular attachment factors.34,36,117 Particle heterogeneity also promotes the exposure of the viral membrane as a prerequisite for using apoptotic mimicry in virus entry,41 a mechanism that has yet to be investigated for TBEV. Populations of heterogeneous particles may not only be essential for virus replication in quite distantly related invertebrate and vertebrate hosts, but also modulate antibody responses and epitope recognition.109,110,118 These are new exciting aspects of flavivirus structure that provide a fertile ground for interesting and important investigations in the future, aiming at a more profound understanding of the complex biology of TBEV as a human pathogen.
Acknowledgments
Research performed by the authors was supported by the Austrian Science Fund FWF.
Contact:
franz.x.heinz@meduniwien.ac.at
Citation:
Heinz FX, Stiasny K. The molecular and antigenic structure of TBEV. Chapter 2b. In: Dobler G, Erber W, Bröker M, Schmitt HJ, eds. The TBE Book. 6th ed. Singapore: Global Health Press; 2023. doi: 10.33442/26613980_2b-6
References
- Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature. 1995;375(6529):291-8.
- Modis Y, Ogata S, Clements D, Harrison SC. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc Natl Acad Sci U S A. 2003;100(12):6986-91.
- Modis Y, Ogata S, Clements D, Harrison SC. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J Virol. 2005;79(2):1223-31.
- Kanai R, Kar K, Anthony K, Gould LH, Ledizet M, Fikrig E, et al. Crystal Structure of West Nile Virus Envelope Glycoprotein Reveals Viral Surface Epitopes. J Virol. 2006;80(22):11000-8.
- Zhang Y, Zhang W, Ogata S, Clements D, Strauss JH, Baker TS, et al. Conformational changes of the flavivirus E glycoprotein. Structure. 2004;12(9):1607-18.
- Nybakken GE, Nelson CA, Chen BR, Diamond MS, Fremont DH. Crystal Structure of the West Nile Virus Envelope Glycoprotein. J Virol. 2006;80(23):11467-74.
- Luca VC, AbiMansour J, Nelson CA, Fremont DH. Crystal structure of the Japanese encephalitis virus envelope protein. J Virol. 2012;86(4):2337-46.
- Barba-Spaeth G, Dejnirattisai W, Rouvinski A, Vaney M-C, Medits I, Sharma A, et al. Structural basis of potent Zika–dengue virus antibody cross-neutralization. 2016;536(7614):48-53.
- Klitting R, Roth L, Rey FA, de Lamballerie X. Molecular determinants of Yellow Fever Virus pathogenicity in Syrian Golden Hamsters: one mutation away from virulence. Emerg Microbes Infect. 2018;7(1):51.
- Dai L, Song J, Lu X, Deng YQ, Musyoki AM, Cheng H, et al. Structures of the Zika Virus Envelope Protein and Its Complex with a Flavivirus Broadly Protective Antibody. Cell Host Microbe. 2016;19(5):696-704.
- Lu X, Xiao H, Li S, Pang X, Song J, Liu S, et al. Double lock of a human neutralizing and protective monoclonal antibody targeting the yellow fever virus envelope. Cell Rep. 2019;26(2):438-46.e5.
- Simmonds P, Becher P, Collett MS, Gould EA, Heinz FX, Meyers G, et al. Family Flaviviridae. In: King AMQ, Lefkowitz E, Adams MJ, Carstens EB, editors. Virus Taxonomy IXth Report of the International Committee on Taxonomy of Viruses. San Diego: Elsevier Academic Press; 2011. p. 1003-20.
- Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, et al. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108(5):717-25.
- Mukhopadhyay S, Kim BS, Chipman PR, Rossmann MG, Kuhn RJ. Structure of West Nile virus. Science (New York, NY. 2003;302(5643):248.
- Zhang W, Chipman PR, Corver J, Johnson PR, Zhang Y, Mukhopadhyay S, et al. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nature Struct Biol. 2003;10(11):907-12.
- Zhang Y, Corver J, Chipman PR, Zhang W, Pletnev SV, Sedlak D, et al. Structures of immature flavivirus particles. EMBO J. 2003;22(11):2604-13.
- Zhang W, Kaufmann B, Chipman PR, Kuhn RJ, Rossmann MG. Membrane curvature in flaviviruses. J Struct Biol. 2013;183(1):86-94.
- Sirohi D, Chen Z, Sun L, Klose T, Pierson TC, Rossmann MG, et al. The 3.8 Å resolution cryo-EM structure of Zika virus. Science. 2016;352(6284):467-70.
- Prasad VM, Miller AS, Klose T, Sirohi D, Buda G, Jiang W, et al. Structure of the immature Zika virus at 9 A resolution. Nat Struct Mol Biol. 2017;24(2):184-6.
- Sevvana M, Long F, Miller AS, Klose T, Buda G, Sun L, et al. Refinement and analysis of the mature Zika virus cryo-EM structure at 3.1Å resolution. 2018;26(9):1169-77.e3.
- Kostyuchenko VA, Zhang Q, Tan JL, Ng TS, Lok SM. Immature and mature dengue serotype 1 virus structures provide insight into the maturation process. J Virol. 2013;87(13):7700-7.
- Kostyuchenko VA, Chew PL, Ng TS, Lok SM. Near-atomic resolution cryo-electron microscopic structure of dengue serotype 4 virus. J Virol. 2014;88(1):477-82.
- Kostyuchenko VA, Lim EXY, Zhang S, Fibriansah G, Ng T-S, Ooi JSG, et al. Structure of the thermally stable Zika virus. Nature. 2016;533(7603):425-8.
- Zhang X, Ge P, Yu X, Brannan JM, Bi G, Zhang Q, et al. Cryo-EM structure of the mature dengue virus at 3.5-A resolution. Nat Struct Mol Biol. 2013;20(1):105-10.
- Wang X, Li S-H, Zhu L, Nian Q-G, Yuan S, Gao Q, et al. Near-atomic structure of Japanese encephalitis virus reveals critical determinants of virulence and stability. Nature Commun. 2017;8:14.
- Füzik T, Formanová P, Růžek D, Yoshii K, Niedrig M, Plevka P. Structure of tick-borne encephalitis virus and its neutralization by a monoclonal antibody. Nature Commun. 2018;9(1):436.
- Lindenbach BD, Murray CL, Thiel HJ, Rice CM. Flaviviridae. In: Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, et al., editors. Fields Virology. 6 ed. Philadelphia: Lippincott. Williams & Wilkins.; 2013. p. 712-46.
- Lorenz IC, Allison SL, Heinz FX, Helenius A. Folding and dimerization of tick-borne encephalitis virus envelope proteins prM and E in the endoplasmic reticulum. J Virol. 2002;76(11):5480-91.
- Heinz FX, Stiasny K, Puschner-Auer G, Holzmann H, Allison SL, Mandl CW, et al. Structural changes and functional control of the tick-borne encephalitis virus glycoprotein E by the heterodimeric association with protein prM. Virology. 1994;198(1):109-17.
- Stadler K, Allison SL, Schalich J, Heinz FX. Proteolytic activation of tick-borne encephalitis virus by furin. J Virol. 1997;71(11):8475-81.
- Yu IM, Zhang W, Holdaway HA, Li L, Kostyuchenko VA, Chipman PR, et al. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science 2008;319(5871):1834-7.
- Elshuber S, Allison SL, Heinz FX, Mandl CW. Cleavage of protein prM is necessary for infection of BHK-21 cells by tick-borne encephalitis virus. J Gen Virol. 2003;84(Pt 1):183-91.
- Yu IM, Holdaway HA, Chipman PR, Kuhn RJ, Rossmann MG, Chen J. Association of the pr peptides with dengue virus at acidic pH blocks membrane fusion. J Virol. 2009;83(23):12101-7.
- Rey FA, Stiasny K, Heinz FX. Flavivirus structural heterogeneity: implications for cell entry. Current Opin Virol. 2017;24:132-9.
- Rey FA, Stiasny K, Vaney MC, Dellarole M, Heinz FX. The bright and the dark side of human antibody responses to flaviviruses: lessons for vaccine design. EMBO reports. 2018;19(2):206-24.
- Kuhn RJ, Dowd KA, Beth Post C, Pierson TC. Shake, rattle, and roll: Impact of the dynamics of flavivirus particles on their interactions with the host. Virology. 2015;479-480C:508-17.
- Heinz FX, Mandl C, Berger R, Tuma W, Kunz C. Antibody-induced conformational changes result in enhanced avidity of antibodies to different antigenic sites on the tick-borne encephalitis virus glycoprotein. Virology. 1984;133(1):25-34.
- Cherrier MV, Kaufmann B, Nybakken GE, Lok SM, Warren JT, Chen BR, et al. Structural basis for the preferential recognition of immature flaviviruses by a fusion-loop antibody. EMBO J. 2009;28(20):3269-76.
- Plevka P, Battisti AJ, Junjhon J, Winkler DC, Holdaway HA, Keelapang P, et al. Maturation of flaviviruses starts from one or more icosahedrally independent nucleation centres. EMBO Rep. 2011;12(6):602-6.
- Perera-Lecoin M, Meertens L, Carnec X, Amara A. Flavivirus entry receptors: an update. Viruses. 2014;6(1):69-88.
- Amara A, Mercer J. Viral apoptotic mimicry. Nat Rev Micro. 2015;13(8):461-9.
- Lessler J, Chaisson LH, Kucirka LM, Bi Q, Grantz K, Salje H, et al. Assessing the global threat from Zika virus. Science. 2016;353(6300):aaf8160.
- Weaver SC. Emergence of Epidemic Zika Virus Transmission and Congenital Zika Syndrome: Are Recently Evolved Traits to Blame? mBio. 2017;8(1):e02063-16.
- Russell P, Brandt W, Dalrymple J. Chemical and antigenic structure of flaviviruses. The togaviruses. Academic Press, New York, NY. 1980; pp. 503-29.
- Westaway EG. Flavivirus replication strategy. Adv Virus Res. 1987;33:45-90.
- Allison SL, Mandl CW, Kunz C, Heinz FX. Expression of cloned envelope protein genes from the flavivirus tick-borne encephalitis virus in mammalian cells and random mutagenesis by PCR. Virus Genes. 1994;8(3):187-98.
- Allison SL, Stadler K, Mandl CW, Kunz C, Heinz FX. Synthesis and secretion of recombinant tick-borne encephalitis virus protein E in soluble and particulate form. J Virol. 1995;69(9):5816-20.
- Schalich J, Allison SL, Stiasny K, Mandl CW, Kunz C, Heinz FX. Recombinant subviral particles from tick-borne encephalitis virus are fusogenic and provide a model system for studying flavivirus envelope glycoprotein functions. J Virol. 1996;70(7):4549-57.
- Blazevic J, Rouha H, Bradt V, Heinz FX, Stiasny K. Membrane Anchors of the Structural Flavivirus Proteins and Their Role in Virus Assembly. J Virol. 2016;90(14):6365-78.
- Allison SL, Stiasny K, Stadler K, Mandl CW, Heinz FX. Mapping of functional elements in the stem-anchor region of tick-borne encephalitis virus envelope protein E. J Virol. 1999;73(7):5605-12.
- Lorenz IC, Kartenbeck J, Mezzacasa A, Allison SL, Heinz FX, Helenius A. Intracellular assembly and secretion of recombinant subviral particles from tick-borne encephalitis virus. J Virol. 2003;77(7):4370-82.
- Ferlenghi I, Clarke M, Ruttan T, Allison SL, Schalich J, Heinz FX, et al. Molecular organization of a recombinant subviral particle from tick-borne encephalitis virus. Mol Cell. 2001;7(3):593-602.
- Allison SL, Tao YJ, O’Riordain G, Mandl CW, Harrison SC, Heinz FX. Two distinct size classes of immature and mature subviral particles from tick-borne encephalitis virus. J Virol. 2003;77(21):11357-66.
- Corver J, Ortiz A, Allison SL, Schalich J, Heinz FX, Wilschut J. Membrane fusion activity of tick-borne encephalitis virus and recombinant subviral particles in a liposomal model system. 2000;269(1):37-46.
- Allison SL, Schalich J, Stiasny K, Mandl CW, Heinz FX. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J Virol. 2001;75(9):4268-75.
- Stiasny K, Kiermayr S, Holzmann H, Heinz FX. Cryptic properties of a cluster of dominant flavivirus cross-reactive antigenic sites. J Virol. 2006;80(19):9557-68.
- Fritz R, Stiasny K, Heinz FX. Identification of specific histidines as pH sensors in flavivirus membrane fusion. J Cell Biol. 2008;183(2):353-61.
- Kiermayr S, Stiasny K, Heinz FX. Impact of quaternary organization on the antigenic structure of the tick-borne encephalitis virus envelope glycoprotein E. J Virol. 2009;83(17):8482-91.
- Fritz R, Blazevic J, Taucher C, Pangerl K, Heinz FX, Stiasny K. The unique transmembrane hairpin of flavivirus fusion protein E is essential for membrane fusion. J Virol. 2011;85(9):4377-85.
- Pangerl K, Heinz FX, Stiasny K. Mutational analysis of the zippering reaction during flavivirus membrane fusion. J Virol. 2011;85(17):8495-501.
- Heinz FX, Allison SL, Stiasny K, Schalich J, Holzmann H, Mandl CW, et al. Recombinant and virion-derived soluble and particulate immunogens for vaccination against tick-borne encephalitis. Vaccine. 1995;13(17):1636-42.
- Aberle JH, Aberle SW, Allison SL, Stiasny K, Ecker M, Mandl CW, et al. A DNA immunization model study with constructs expressing the tick-borne encephalitis virus envelope protein E in different physical forms. J Immunol. 1999;163(12):6756-61.
- Mandl CW, Kroschewski H, Allison SL, Kofler R, Holzmann H, Meixner T, et al. Adaptation of tick-borne encephalitis virus to BHK-21 cells results in the formation of multiple heparan sulfate binding sites in the envelope protein and attenuation in vivo. J Virol. 2001;75(12):5627-37.
- Lee E, Lobigs M. Substitutions at the Putative Receptor-Binding Site of an Encephalitic Flavivirus Alter Virulence and Host Cell Tropism and Reveal a Role for Glycosaminoglycans in Entry. J Virol. 2000;74(19):8867-75.
- Lee E, Lobigs M. Mechanism of Virulence Attenuation of Glycosaminoglycan-Binding Variants of Japanese Encephalitis Virus and Murray Valley Encephalitis Virus. J Virol. 2002;76(10):4901-11.
- Lee E, Hall RA, Lobigs M. Common E Protein Determinants for Attenuation of Glycosaminoglycan-Binding Variants of Japanese Encephalitis and West Nile Viruses. J Virol. 2004;78(15):8271-80.
- Lee E, Lobigs M. E Protein Domain III Determinants of Yellow Fever Virus 17D Vaccine Strain Enhance Binding to Glycosaminoglycans, Impede Virus Spread, and Attenuate Virulence. J Virol. 2008;82(12):6024-33.
- Kroschewski H, Allison SL, Heinz FX, Mandl CW. Role of heparan sulfate for attachment and entry of tick-borne encephalitis virus. 2003;308(1):92-100.
- Stuart D, Gouet P. Viral envelope glycoproteins swing into action. 1995;3(7):645-8.
- Anderson R. Manipulation of cell surface macromolecules by flaviviruses. Advances in Virus Research. Volume 59: Academic Press; 2003. pp. 229-74.
- Ruoslahti E. RGD and other recognition sequences for integrins. Annu Rev Cell Dev Biol. 1996;12:697-715.
- Bhardwaj S, Holbrook M, Shope RE, Barrett ADT, Watowich SJ. Biophysical Characterization and Vector-Specific Antagonist Activity of Domain III of the Tick-Borne Flavivirus Envelope Protein. J Virol. 2001;75(8):4002-7.
- Meertens L, Carnec X, Lecoin MP, Ramdasi R, Guivel-Benhassine F, Lew E, et al. The TIM and TAM families of phosphatidylserine receptors mediate dengue virus entry. Cell Host Microbe. 2012;12(4):544-57.
- Hamel R, Dejarnac O, Wichit S, Ekchariyawat P, Neyret A, Luplertlop N, et al. Biology of Zika Virus Infection in Human Skin Cells. J Virol. 2015;89(17):8880-96.
- Ravichandran Kodi. Beginnings of a good apoptotic meal: The find-me and eat-me signaling pathways. 2011;35(4):445-55.
- Segawa K, Nagata S. An apoptotic ‘eat me’ signal: phosphatidylserine exposure. Trends Cell Biol. 2015;25(11):639-50.
- Guirakhoo F, Heinz FX, Mandl CW, Holzmann H, Kunz C. Fusion activity of flaviviruses: comparison of mature and immature (prM-containing) tick-borne encephalitis virions. J Gen Virol. 1991;72:1323-9.
- Stiasny K, Allison SL, Mandl CW, Heinz FX. Role of metastability and acidic pH in membrane fusion by tick-borne encephalitis virus. J Virol. 2001;75(16):7392-8.
- Stiasny K, Allison SL, Schalich J, Heinz FX. Membrane interactions of the tick-borne encephalitis virus fusion protein E at low pH. J Virol. 2002;76(8):3784-90.
- Stiasny K, Koessl C, Heinz FX. Involvement of lipids in different steps of the flavivirus fusion mechanism. J Virol. 2003;77(14):7856-62.
- Stiasny K, Heinz FX. Effect of membrane curvature-modifying lipids on membrane fusion by tick-borne encephalitis virus. J Virol. 2004;78(16):8536-42.
- Stiasny K, Brandler S, Kossl C, Heinz FX. Probing the Flavivirus Membrane Fusion Mechanism by Using Monoclonal Antibodies. J Virol. 2007;81(20):11526-31.
- Stiasny K, Kossl C, Lepault J, Rey FA, Heinz FX. Characterization of a structural intermediate of flavivirus membrane fusion. PLoS Pathog. 2007;3(2):e20.
- Allison SL, Schalich J, Stiasny K, Mandl CW, Kunz C, Heinz FX. Oligomeric rearrangement of tick-borne encephalitis virus envelope proteins induced by an acidic pH. J Virol. 1995;69(2):695-700.
- Stiasny K, Allison SL, Marchler-Bauer A, Kunz C, Heinz FX. Structural requirements for low-pH-induced rearrangements in the envelope glycoprotein of tick-borne encephalitis virus. J Virol. 1996;70(11):8142-7.
- Stiasny K, Bressanelli S, Lepault J, Rey FA, Heinz FX. Characterization of a membrane-associated trimeric low-pH-induced form of the class II viral fusion protein E from tick-borne encephalitis virus and its crystallization. J Virol. 2004;78(6):3178-83.
- Bressanelli S, Stiasny K, Allison SL, Stura EA, Duquerroy S, Lescar J, et al. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 2004;23(4):728-38.
- Kreil TR, Maier E, Fraiss S, Eibl MM. Neutralizing antibodies protect against lethal flavivirus challenge but allow for the development of active humoral immunity to a nonstructural virus protein. Virol. 1998;72(4):3076-81.
- Heinz FX, Tuma W, Guirakhoo F, Berger R, Kunz C. Immunogenicity of tick-borne encephalitis virus glycoprotein fragments: epitope-specific analysis of the antibody response. J Gen Virol. 1984;65:1921-9.
- Heinz FX, Berger R, Majdic O, Knapp W, Kunz C. Monoclonal antibodies to the structural glycoprotein of tick-borne encephalitis virus. Infect Immun. 1982;37(3):869-74.
- Guirakhoo F, Heinz FX, Kunz C. Epitope model of tick-borne encephalitis virus envelope glycoprotein E: analysis of structural properties, role of carbohydrate side chain, and conformational changes occurring at acidic pH. Virology. 1989;169(1):90-9.
- Holzmann H, Stiasny K, York H, Dorner F, Kunz C, Heinz FX. Tick-borne encephalitis virus envelope protein E-specific monoclonal antibodies for the study of low pH-induced conformational changes and immature virions. Arch Virol. 1995;140(2):213-21.
- Heinz FX, Berger R, Tuma W, Kunz C. A topological and functional model of epitopes on the structural glycoprotein of tick-borne encephalitis virus defined by monoclonal antibodies. 1983;126(2):525-37.
- Holzmann H, Mandl CW, Guirakhoo F, Heinz FX, Kunz C. Characterization of antigenic variants of tick-borne encephalitis virus selected with neutralizing monoclonal antibodies. J Gen Virol. 1989;70:219-22.
- Mandl CW, Guirakhoo F, Holzmann H, Heinz FX, Kunz C. Antigenic structure of the flavivirus envelope protein E at the molecular level, using tick-borne encephalitis virus as a model. J Virol. 1989;63(2):564-71.
- Holzmann H, Stiasny K, Ecker M, Kunz C, Heinz FX. Characterization of monoclonal antibody-escape mutants of tick-borne encephalitis virus with reduced neuroinvasiveness in mice. J Gen Virol. 1997;78:31-7.
- Heinz FX, Stiasny K. Flaviviruses and their antigenic structure. J Clin Virol. 2012;55(4):289-95.
- Dowd KA, Pierson TC. Antibody-mediated neutralization of flaviviruses: a reductionist view. 2011;411(2):306-15.
- Lok S-M. The interplay of dengue virus morphological diversity and human antibodies. Trends Microbiol. 2016;24(4):284-93.
- Diamond Michael, Pierson Theodore. Molecular Insight into Dengue Virus Pathogenesis and Its Implications for Disease Control. Cell. 2015;162(3):488-92.
- Pierson TC, Diamond MS. Flaviviruses. In: Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, et al., eds. Fields Virology. 6 ed. Philadelphia: Lippincott. Williams & Wilkins; 2013. pp. 747-94.
- Pierson TC, Fremont DH, Kuhn RJ, Diamond MS. Structural insights into the mechanisms of antibody-mediated neutralization of flavivirus infection: implications for vaccine development. Cell Host Microbe. 2008;4(3):229-38.
- Screaton G, Mongkolsapaya J, Yacoub S, Roberts C. New insights into the immunopathology and control of dengue virus infection. Nat Rev Immunol. 2015;15(12):745-59.
- Demina TV, Dzhioev YP, Verkhozina MM, Kozlova IV, Tkachev SE, Plyusnin A, et al. Genotyping and characterization of the geographical distribution of tick-borne encephalitis virus variants with a set of molecular probes. J Med Virol. 2010;82(6):965-76.
- Chernokhaeva LL, Rogova YV, Vorovitch MF, Romanova LI, Kozlovskaya LI, Maikova GB, et al. Protective immunity spectrum induced by immunization with a vaccine from the TBEV strain Sofjin. Vaccine. 2016;34(20):2354-61.
- Kovalev SY, Mukhacheva TA. Tick-borne encephalitis virus subtypes emerged through rapid vector switches rather than gradual evolution. Ecol Evol. 2014;4(22):4307-16.
- Orlinger KK, Hofmeister Y, Fritz R, Holzer GW, Falkner FG, Unger B, et al. A Tick-borne Encephalitis Virus Vaccine Based on the European Prototype Strain Induces Broadly Reactive Cross-neutralizing Antibodies in Humans. J Infect Dis. 2011;203(11):1556-64.
- Fritz R, Orlinger KK, Hofmeister Y, Janecki K, Traweger A, Perez-Burgos L, et al. Quantitative comparison of the cross-protection induced by tick-borne encephalitis virus vaccines based on European and Far Eastern virus subtypes. Vaccine. 2012;30(6):1165-9.
- Dowd KA, DeMaso CR, Pierson TC. Genotypic Differences in Dengue Virus Neutralization Are Explained by a Single Amino Acid Mutation That Modulates Virus Breathing. 2015;6(6):e01559-15.
- Goo L, VanBlargan LA, Dowd KA, Diamond MS, Pierson TC. A single mutation in the envelope protein modulates flavivirus antigenicity, stability, and pathogenesis. PLoS Pathog. 2017;13(2):e1006178.
- Beck Y, Fritz R, Orlinger K, Kiermayr S, Ilk R, Portsmouth D, et al. Molecular Basis of the Divergent Immunogenicity of Two Pediatric Tick-Borne Encephalitis Virus Vaccines. J Virol 2016;90(4):1964-72.
- Jarmer J, Zlatkovic J, Tsouchnikas G, Vratskikh O, Strauss J, Aberle JH, et al. Variation of the specificity of the human antibody responses after tick-borne encephalitis virus infection and vaccination. J Virol. 2014;88(23):13845-57.
- Tsouchnikas G, Zlatkovic J, Jarmer J, Strauss J, Vratskikh O, Kundi M, et al. Immunization with immune complexes modulates the fine specificity of antibody responses to a flavivirus antigen. J Virol. 2015;89(15):7970-8.
- Mukhopadhyay S, Kuhn RJ, Rossmann MG. A structural perspective of the flavivirus life cycle. Nat Rev Microbiol. 2005;3(1):13-22.
- Kaufmann B, Rossmann MG. Molecular mechanisms involved in the early steps of flavivirus cell entry. Microb Infect. 2011;13(1):1-9.
- Pierson TC, Diamond MS. Degrees of maturity: the complex structure and biology of flaviviruses. Curr Opin Virol. 2012;2(2):168-75.
- Dowd KA, Pierson TC. The many faces of a dynamic virion: implications of viral breathing on flavivirus biology and immunogenicity. Annu Rev Virol. 2018;5(1):185-207.
- Dowd KA, Mukherjee S, Kuhn RJ, Pierson TC. Combined effects of the structural heterogeneity and dynamics of flaviviruses on antibody recognition. J Virol. 2014;88(20):11726-37.
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605-12